RETROGEN SEQUENCING TROUBLESHOOTING GUIDE AND SUGGESTIONS
I. PREPARATION OF DNA TEMPLATE
The quality of the DNA template is one of the most critical factors in DNA sequencing. Please make certain that the DNA template is at the proper concentration, free of contaminants, dissolved in distilled water, and that the DNA is not degraded.
ISOLATING THE DNA
Plasmids: For plasmid preparations, numerous commercial protocols work in addition to traditional methods. It is essential to avoid genomic DNA, excess RNA, and contaminants. Elute in distilled water or in 1 mM Tris. Do not elute in TE, as the EDTA can interfere with sequencing. Cesium Chloride preps work, but be certain there are no Cs or EDTA present in the final sample. An extra ethanol precipitation is helpful to eliminate these contaminants.
PCR Products: If there is a single amplification, a commercial column that separates the PCR products from primers and enzymes can be used, or an enzymatic cleanup reaction can be done to remove all single-stranded DNA from the sample. Alternately, when multiple regions are amplified, the PCR products can be electrophoresed and the desired band can be excised and gel-eluted. Extraneous bands may appear low-intensity, but they could easily contaminate the sequencing run. It is essential to remove all traces of the original PCR primers, as these could produce undesired sequencing products by acting as sequencing primers (recall that PCR is a bi-directional exponential reaction, whereas sequencing is a uni-directional linear reaction).
Large Clones (BAC, Cosmid, etc.): BAC (and other) preps are commonly contaminated with 20-80% genomic DNA. This excess DNA decreases the effective concentration of the template, so genomic DNA must be avoided. It is imperative that gentle handling of the samples is maintained during the alkaline lysis step. During the potassium acetate precipitation step to remove protein/genomic DNA, extremely gentle handling is required. If you are having difficulty sequencing these types of samples, perform a restriction digest of the DNA and examine the bands on a gel. If any smear in the background is visible, there is too much genomic DNA in the prep.
QUANTITATION OF THE TEMPLATE DNA:
Provided there is ample template DNA, it is best to determine the concentration by UV absorption. For success in getting accurate spec readings on DNA, please consider the following points:
Readings below 0.05 AU require careful blanking in order to ensure accuracy and many specs are unreliable below 0.05 AU.
Contaminants such as RNA and free nucleotides also absorb UV, affecting the AU value.
Avoid chromosomal/genomic DNA contamination. Upon restriction digestion, genomic DNA contamination is visible as a smear in the background of the lane on a given gel. Any visible smear represents a large percentage of the total DNA present, thus resulting in an artificially inflated UV reading with respect to the template to be sequenced.
If there is not enough DNA to measure spectrophotometrically, then estimate the amount of DNA from a gel by comparing it to known standards. It is good practice to do this even in addition to a spec measurement to verify the result. Further, if the DNA concentration is unable to be accurately determined, good laboratory practice dictates not proceeding until there is enough DNA to measure accurately.
III. DILUTING THE TEMPLATE TO THE DESIRED CONCENTRATION:
After successfully determining the template DNA concentration, dilute the template to the final concentration using distilled water. Do not use TE or other EDTA-containing buffers and do not add any divalent cations (i.e. Mg, Ca, Mn) or salts.
Premixed Service: DNA and Primer Combined
| DNA | AMOUNT | PRIMER | TOTAL VOL. | 2X DNA | 2X PRIMER | 2X TOTAL |
| PCR Product (per KB) | 60ng | 10pmol | 6μL | 120ng | 20pmol | 12μL |
| Single-stranded Plasmid | 150ng | 10pmol | 6μL | 300ng | 20pmol | 12μL |
| Double-stranded Plasmid | 300ng | 10pmol | 6μL | 600ng | 20pmol | 12μL |
| BAC | 2000ng | 10pmol | 6μL | 4000ng | 20pmol | 12μL |
We recommend 2x amount of DNA in case we need to repeat the reactions.
Standard Service: DNA and Primer Separated
DNA | CONCENTRATION | AMOUNT | 2X AMOUNT |
PCR Product (per KB) | DNA 10‐40ng/μL | 50ng | 100ng |
Single-stranded | Plasmid 50‐100ng/μL | 150ng | 300ng |
Double-stranded | Plasmid 50‐200ng/μL | 200ng | 400ng |
BAC | 100‐200ng/μL | 2000ng | 4000ng |
Primer | 5pmol/μL | 3μL | 6μL |
We recommend 2x amount of DNA in case we need to repeat the reactions.
Large Clones (BAC): For large clones, like BAC clones, large amounts of non-template DNA are present. For this reason, the template concentration must be very high since only a small percentage of the clone will be used as sequencing template. To address this issue, the total concentration of DNA must be increased. The best success is achieved with 0.5-1.0μg/μL for these size clones. BAC samples should be visibly viscous and difficult to pipette at this concentration. The excess DNA in the reaction will bind primer non-specifically, so the primer concentration must be increased accordingly. We suggest 10μM as a good standard primer concentration.
IV. PRIMERS:
Special consideration must be given to primers and primer design. For PCR, two primers replicate opposite strands which will quickly outnumber the original template DNA. Most PCR amplifications go beyond the linear part of the reaction, meaning inefficient primers can produce as much product as efficient ones. PCR will amplify the target DNA even if it is only a small proportion of the DNA present. For sequencing, inefficient primers will produce weaker bands and a resulting poor signal. The best sequencing primers are efficient at binding to the target site, have little to no secondary binding site issues, do not self-anneal (primer-dimer formation) or have internal hairpins or secondary structure problems, and have a reasonable G/C to A/T ratio. Please be certain that the sequencing primers meet these minimal standards to ensure a positive sequencing result.
V. TROUBLESHOOTING – EXAMPLES & SOLUTIONS:
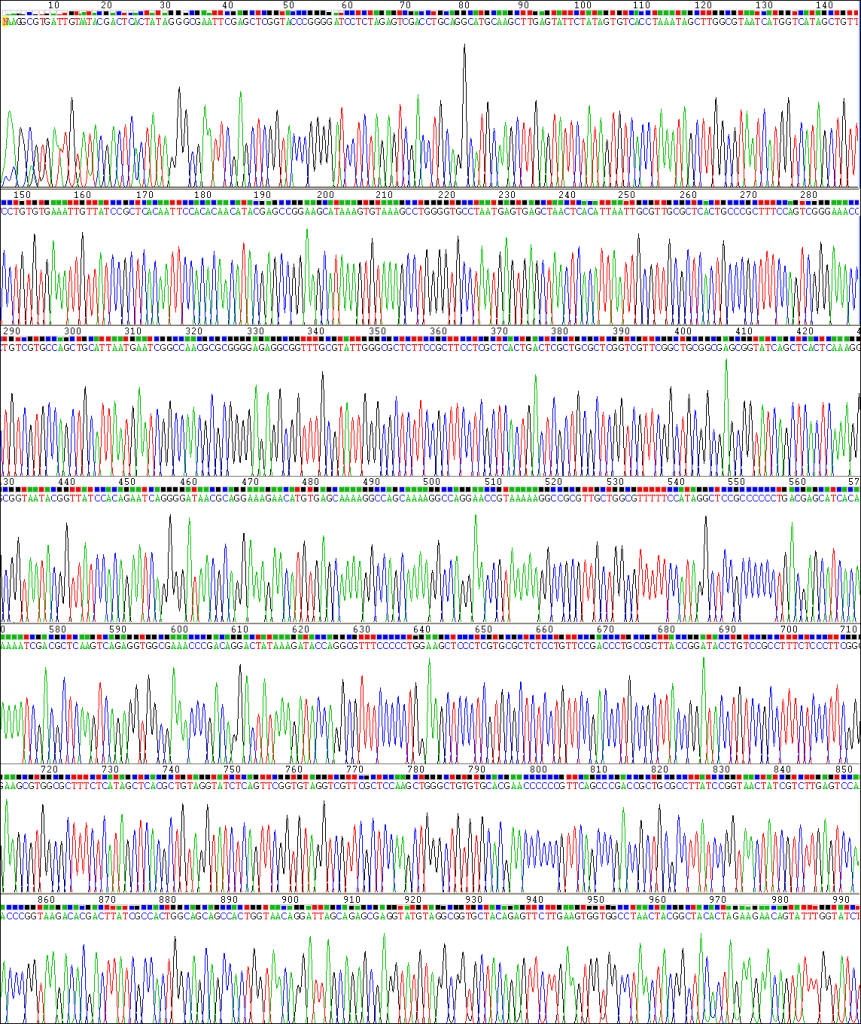
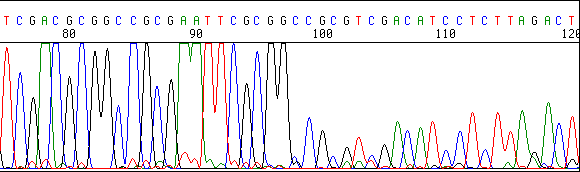
Below is an example of an exceptional-quality sequencing reaction. The peaks are evenly spaced, uniform, and clean. The automatic base-caller will make few, if any, mistakes, providing confidence that the sequence is correct.
This next section discusses troubleshooting examples and solutions of DNA sequencing. When reactions are completed and results are available, an automatic email message is generated and sent to the designated recipient. A follow-up email from our technical staff may be generated for problematic or difficult results which provides troubleshooting suggestions and advice.
The follow-up email from our technical staff may address specific issues regarding the samples such as “weak signal”, “no signal”, “multiple signal” or “short signal.” These results most often reflect difficulties with the template or primer preparations and concentrations. The remainder of this section breaks down the common troubleshooting issues that occur.
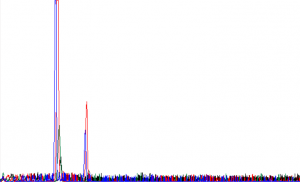
A. WEAK or NO SIGNAL:
In the chromatogram below, the peaks are ill-defined and not much higher than background noise. This is characterized as “weak signal” or “no signal.”
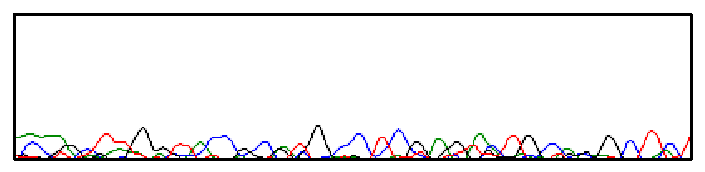
Our positive control reactions have an approximate average signal strength of 1500. Reactions ideally have an average signal strength between 800-1500, although it is possible to have an excellent-looking sequence with signal strengths much higher or lower. However, signals below 200 or above 4000 often have serious problems.
The most common poor result in sequencing is “weak” or “no” signal. DNA preparation is the first consideration. Contaminated templates, low DNA concentrations, unclean samples, and mixed samples can cause this result. The primer design and concentration also factor heavily in the success of the reaction. Some examples and suggestions follow:
DNA is low concentration:
How was the DNA prepared? Has this method of preparation led to successful sequencing reactions in the past? Has anything changed? How was the concentration measured?
Was the DNA concentration estimated from a gel? When sequencing PCR reactions, it is recommended to use an analytical gel to estimate DNA concentration. Gel elution of restriction fragments often must be measured the same way. Please be sure to run the gel on the same tube of DNA you are sending us. Gel elution of fragments does not result in 100% recovery.
Primer concentration/design problems:
There was insufficient primer in the primer tube. Please double-check all calculations. The primer concentrations are specified in pmol/μL – that is picomoles per microliter, not ‘picomolar’. There are six orders of magnitude between them. We prefer a 10μM solution of primer in a clearly labeled tube.
The primer did not interact efficiently with the template. It is recommended that a complete restriction map is generated. This can easily explain cloning issues with regard to the presence, absence, or damage of the priming site.
Did you design the primer from accurate sequence information? Make sure the region used is an accurate read with high-quality base-calling. Common primer design software is available both commercially and academically.
Does the primer function at our annealing temperatures? Since we handle so many samples, we must process them all with the same cycling conditions. We anneal at 50°C.
B. SHORT READS:
Short signals can appear in various ways:
Signals can drop off quickly:
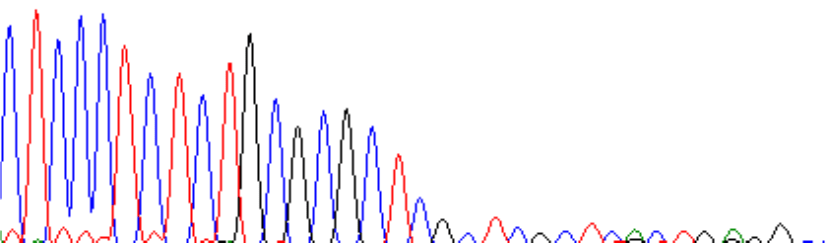
There can be a very gradual signal loss (top heavy data):
At the start:
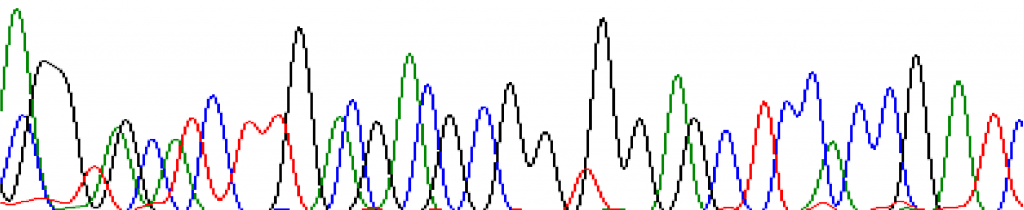
At 200bps:
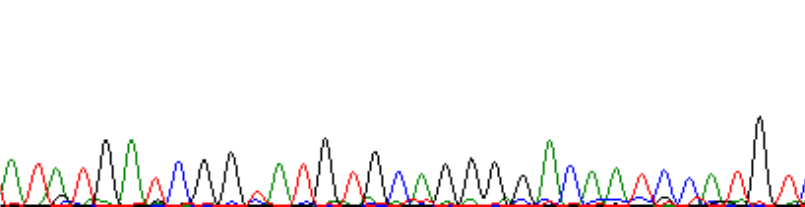
At 400bps:
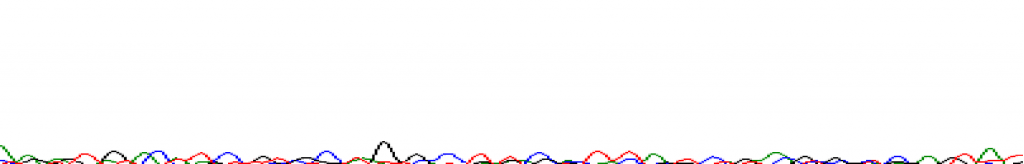
There can be an abrupt drop off:
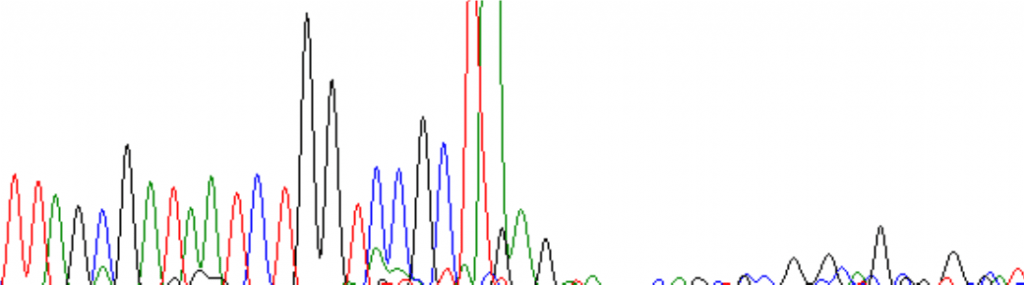
Or there can be a drop off at a GC-rich region:
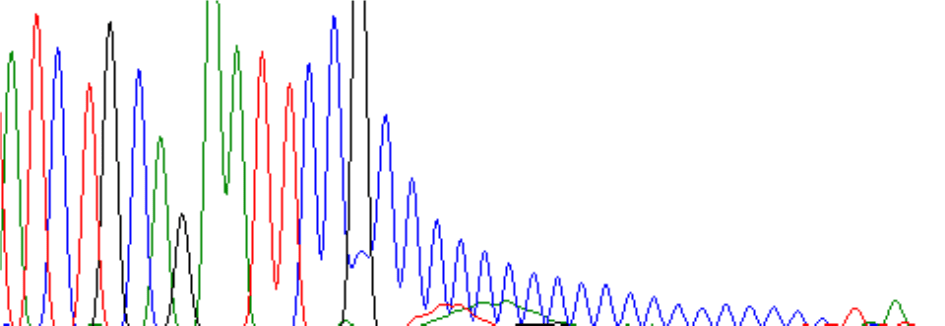
Make sure you inform Retrogen if any template is known to be GC-rich so that we can use optimal conditions for these templates. If these conditions still give short signals, contact Retrogen to discuss further methods for improving results.
There are four known causes for the loss of resolution (LOR) problem. Three of the failure modes are within the sequencing facility itself and are relatively uncommon. We watch for them all the time. The known causes of the LOR problem are as follows:
Air bubbles in the electrophoresis capillaries. If a capillary is partially blocked by an air bubble, the sample in that capillary will exhibit extremely poor resolution (if it elutes at all). This results in sporadic failures of individual samples out of sets that otherwise gave good results. Retrogen will repeat samples exhibiting sporadic LOR upon request.
Plugged cuvette flow ports. The flow paths in the instrument can become blocked, in which case an entire set of samples will fail with loss of resolution – every sample, including standards.
Overloaded lanes. Excessive amounts of sample (sequencing termination products) in a capillary can cause a loss of resolution due to migration issues – see section E for more details.
The last failure mode is very common and is characteristic of entire sets of samples. In other words, it is a sample prep problem.
Contaminated samples. We have time and time again seen entire sets of samples produce LOR when the rest of the samples in that set (including standards) look fine. Samples such as these, when re-run, always give the same LOR result. When the samples are cleaned up (suggestions noted above), they produce excellent results. We can only conclude that there is a contaminant in the samples that reduces resolution in our machines.
Below is an example of a mild ski-slope:
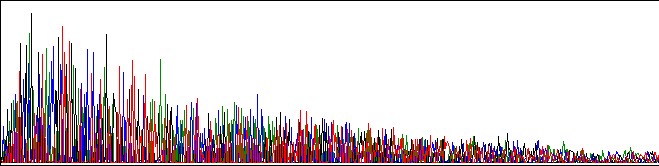
This is not well understood, but here are four possible explanations:
(1) Salt in the DNA.
(2) Too much DNA in the reaction.
(3) An unknown impurity “poisoning” the Taq processivity.
(4) An unknown contaminant increasing the binding of dyes in the enzyme’s active site.
The last effect can arise from free NTPs in the sample, or from a contaminant that disturbs the divalent cation concentration (EDTA, Mg++ , etc.). Buffers such as TE should not be used to resuspend DNA preparations that will be used for sequencing.
Salt is the most common cause of the ‘ski-slope’ effect. Capillary electrophoresis instruments such as our sequencers are quite sensitive to the presence of excess salt. It tends to favor detection of smaller fragments over larger ones. Our purification protocols are designed to minimize this problem, but it still occurs at times.
The terminator concentrations are carefully adjusted to statistically favor long extension and the enzyme is modified to be able to accept bulky dye molecules as substrates. Several of the possible explanations given above for the “ski-slope” effect all work by increasing the statistical likelihood of early termination.
The sequence proceeds normally, then the bands abruptly become much smaller. Secondary structure in the template is the most likely cause of this problem. The polymerase is presumably unable to progress through some stem-loop form. Is it an siRNA (RNAi) construct? These will almost always exhibit strong secondary structure effects. A couple possible solutions are:
1) Try resequencing by selecting ‘siRNA construct’ as your DNA type (best solution).
(2) Try to sequence from another primer at a different position (closer or further).
(3) Sequence the other strand.
We may be able to use special cycling conditions and/or special reagents that help the polymerase to push through this region. We offer additional charged services in this regard, but contact Retrogen to discuss these troubleshooting options.
Here is an example of a secondary structure effect:
C. OTHER TYPES OF CONTAMINANTS
The sequence is generally good, but there is one place where a huge green (or red or black) peak obscures everything under it. The peak shape is clearly abnormal. This is a common artifact of sequencing that arises from complexes formed between the sequencing dyes and other unknown components (often contaminants). If our sample cleanup is flawed, we might have left excess unincorporated dyes in the sample. We will usually catch this, since it is pretty obvious on the array image. Or the sample itself may have a contaminant that binds unincorporated dyes. In either case, you may be able to manually re-call the bases “underneath” the blob-peak. If the Retrogen technicians feel this is not possible, and if the “blob” appears to arise from our own processing problem, we will initiate a no-charge repeat for you upon request.
Here is a typical example of what we call a “dye blob”:
D. MULTIPLE SIGNALS
The first 10-20 nucleotides are obscured by huge, trashy-looking peaks, then normal sequence is seen thereafter. The most likely explanation is that the primer has formed self-dimers and the ‘trash’ peaks are from sequencing on itself. All primers should be designed using a computer in order to avoid such artifacts. Most common primer design programs will avoid primers that form self-dimers.
Alternatively, if the sample is a PCR product, these large peaks may arise from a small PCR product contaminating the main sample. These products are usually not seen on agarose gels since they are such a small amount of the PCR reaction, but they produce signal in a sequencing reaction which makes the data difficult to interpret at best. Cut the PCR product out of the gel to isolate a single band and try again.
The first 20-50 nucleotides are fine, but suddenly the chromatogram shows mixed peaks or terrible background. When the template DNA is actually a mixture of two clones that are identical up to the cloning site and diverge thereafter, multiple sequences are generated. To avoid this problem, always streak out the clones to single colonies to ensure they are completely clonal.
Alternatively, the primer could be sitting down on two independent sites within the construct, generating identical sequence on those two sites up until the point where the two sequences diverge, whereupon you get the peaks-on-peaks effect. This is common when priming inside an insert and there have been two copies of that insert accidentally inserted. Other structural errors can produce this type of effect as well.
Here’s an example of two mixed clones, identical in sequence until they hit the cloning site:
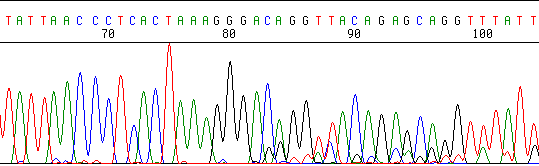
The sequence looks great until it hits a polyA (or polyT) region and then the bands rise and fall in waves. This is called “polymerase slip” or “slippage.” It happens when the growing strand temporarily dissociates from the template, then re-associates at a different spot, usually one nucleotide forward or back from where it started. If this happens often enough (it will on poly-A or poly-T templates), every individual band becomes a family of closely-spaced peaks, giving a ‘roller coaster’ look to the chromatogram. Try sequencing in the other direction from the opposite strand, or try another primer either closer or further from the homopolymer region.
Example of homopolymeric (G-rich) region causing multiple signals:
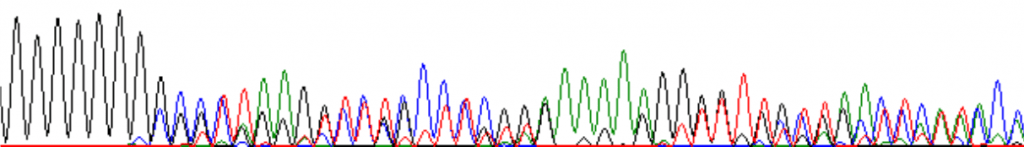
The following is an excellent example of ‘polymerase slip’ on a homopolymeric tract:
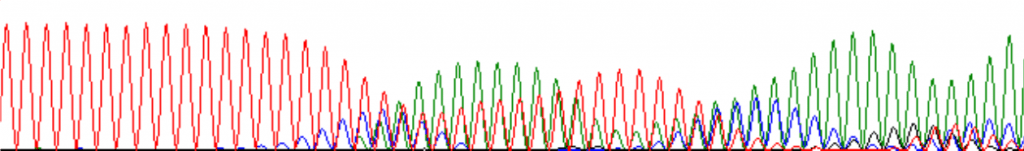
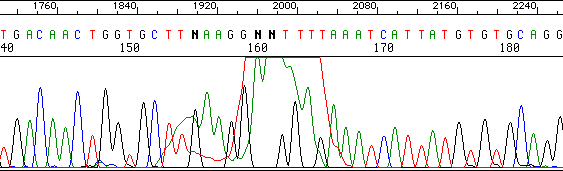
The bands are present and strong, but irregularly spaced, or with mixed colors. This results when two sequences have been superimposed on each other. There are several common causes:
(1) The sequencing primer binds to two (or more) sites on the template.
(2) There are two (or more) templates present.
(3) This was a PCR reaction and the original primers were not completely removed.
(4) This was a PCR reaction and one primer generated both ends of the product.
(5) This was a PCR reaction and there is more than one amplified species present.
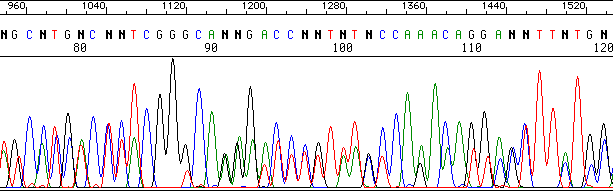
Here is an example of ‘mixed peaks’ that might arise from two or more unrelated templates:
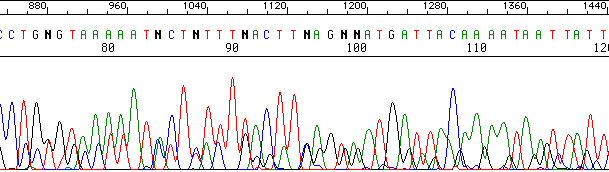
Another example, this time with templates that might be related. Note the alignment of the peaks:
E. MIGRATION ISSUES
Sometimes, there are issues with how the DNA migrates through the capillaries. As a result, the signal could show up late, as shown in the example below:
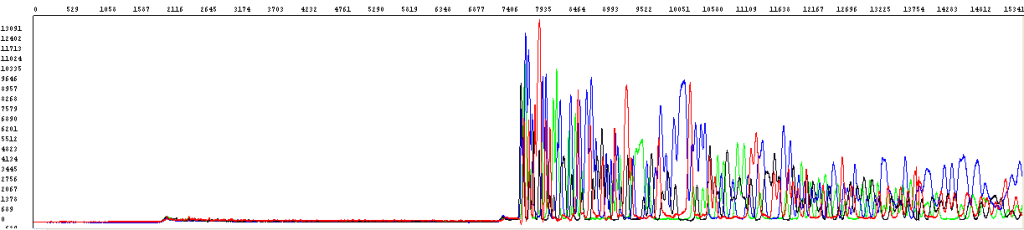
Often times, this is the result of the DNA having a concentration that is too high, thus clogging a particular capillary and causing a delayed signal.
Migration issues can also cause samples to have losses in resolution. Typical examples of this are as follows:
Early in the run, we see reasonable resolution:
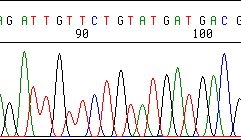
50 or 60 nt further on, it is obvious something is wrong:
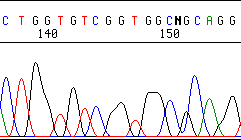
Another 120 nt – the resolution is terrible and the sequence is essentially unreadable:

This loss of resolution is most often a result of a contaminant in the sequencing reaction or the overloading of too much DNA in the sequencing reaction. Some successful suggestions for removing the unknown contaminant are:
Perform a phenol-chloroform extraction on your template DNA.
Perform ammonium acetate-isopropyl precipitation on your template DNA.
Pass your sample through a Sephadex G50 spin column.
Many sequencing facilities have observed that overloaded Qiagen preps (or similar silica preps) are at fault for loss of resolution. When overloaded, these plasmid prep kits leave many impurities in the DNA. To avoid overloading, try at least one of the following:
Grow your minipreps for a shorter time. 12 hours is good; 8 hours is better. Yes, the yield may be reduced, but the DNA sequence template will be better.
Grow smaller bacterial cultures. If the kit says no more than 5 mL of lysate, limit yourself to 3 mL. Yes, the yield will be reduced, but again, the DNA sequence template will be better.
E. FAILED REACTIONS
At Retrogen, we examine virtually every lane from every sample we sequence. We diagnose the problems whenever we can, but some problems just keep coming up again and again. Our sequence analysts spend considerable time with clients who have had problems and carefully examine their experimental design and protocols for the cause of the failure. The following table is a compilation of the most common failure modes seen at Retrogen:
Inadequate template concentration:
Spec readings too low to be accurate
Inexperience at gel estimation of band intensity
Failure to check PCR product concentration
Spec reading with excessive RNA present
Spec reading with excessive genomic DNA present
Overloaded miniprep purification devices (“Loss of Resolution” samples)
Failure to streak clones to single-colony
Calculation error in primer concentration
Multiple priming sites present
Multiple inserts present (gives multiple priming or stem-loop)
Sequencing incorrect PCR products
Failure to fully characterize a new plasmid construct
Secondary structure in clone (siRNA constructs, etc.)
Homopolymer tract (e.g., 3′ sequencing on a cDNA clone)
Salt contamination in sample (“ski-slope” effect, typically gel-eluted frags)
Mis-paired primers (cross-species primer design, inaccurate sequence basis)
Free dNTPs present in template
Primer dimers
Primer Tm too low
Mixed-up samples and/or primers
VI. CONCLUSION:
Here at Retrogen, we strive to provide a clear and long read from the templates submitted. We have spent significant effort to make our services streamlined and successful for the vast majority of templates available. It is true that not all samples fit into this generalized criteria and we are dedicated to resolving such issues with individual clients. Through individual follow-up and customer service, we are able to resolve nearly all sequencing issues and generate quality sequence data. This is not to say that every single sample from every customer results in perfection, but we strive to achieve that standard.
If this guide has not satisfied or answered the troubleshooting requirements from your sample results, please call us at (858) 455-8411 or e-mail us at sequencing@retrogen.com and one of our sequencing analysts will discuss the results, the experimental design, and provide suggestions for getting those positive results.





